7 min read
News Release: Study Identifies Potential COVID Vaccine & Therapy Research Pathways
Conformational change of the spike protein furin cleavage domain may enhance virus infectivity
June 16, 2020 (Santa Clara, Calif.) — Research by two U.S.-based antibody discovery companies shows a rise and dominance of a mutant SARS-CoV-2 virus and identifies a new mechanism predicting a competitive advantage of the mutation. The mutation is predicted to induce a structural change in the spike protein of the virus that would enhance virus entry. The study, conducted by Antibody Solutions and Single Cell Technology (SCT), is available on Preprints.org (the study’s findings have not yet been peer-reviewed).
According to John Kenney, PhD, president and co-founder of Antibody Solutions, advanced molecular modeling may have revealed how D614G, the dominant mutation isolated from a majority of patients in Europe and the eastern U.S., gained a competitive advantage over the earlier isolates of SARS-CoV-2 found in China.
“Our results from gene sequencing clearly show the D614G mutation is more common as the pandemic unfolds,” Dr. Kenney said. “Our homology modeling identifies a mechanism whereby the D614G mutation favors the orientation of critical residues in the furin cleavage domain. You can think of the recently discovered furin domain as an activation sequence or ‘trigger.’ A better orientation of this domain for cleavage would be expected to enhance infectivity.”
The spike protein of SARS-CoV-2 is considered a primary target for COVID-19 therapeutic intervention. Cleavage of the spike protein by furin is a key mechanism distinguishing SARS-CoV-2 from SARS-CoV and non-pathogenic coronaviruses.
“Our findings of the competitive and mechanistic advantages arising from the D614G mutation strengthen the rationale for targeting the furin cleavage domain of the spike protein with vaccines or neutralizing antibodies to inhibit COVID-19 progression,” Dr. Kenney said.
Genetic sequencing confirms geographic distributions of mutations
Chun-nan Chen, PhD, chief science officer and CEO of SCT, used his company’s informatics platform to mine worldwide SARS-CoV-2 genome data to learn more about how the mutants’ efficiency and geography may be correlated. Through genetic sequencing analysis of 11,542 viral genome records from the GISAID database collected through April 28, 2020, Dr. Chen said his team was able to compute the frequency of different mutations.
“By examining amino acid changes in each case, we identified mutations along the Spike, or ‘S,’ protein at 103 positions,” he said, “And D614G was, put simply, the dominant mutation, occurring in 56% of all sequences—far more than all of the other identified mutations combined.
“When we examined the geographical distribution of G614-containing viral genomes,” he continued, “distribution patterns emerged that show the G614 mutant as the dominant SARS-CoV-2 mutant in Europe and large swathes of the U.S. But viral sequences from Chinese and South Korean patients and those from the West Coast of the U.S. were mainly found to be carrying the Wuhan Spike or ‘S’ protein genotype.”
The companies’ research included a comparative analysis of daily cumulative genotype counts by country and by U.S. states, which allowed its investigators to examine which specific viral sequences outpaced others as samples were collected in each geography over time. The most striking observation was that the D614G mutated virus became the predominant form of the virus sampled in most countries and U.S. states where the mutated and non-mutant virus appeared at the same time.
Molecular modeling reveals unexpected changes in D614G structure
That the rise in the D614G mutation is simply the result of a founder effect is possible but unlikely, Dr. Kenney and his team believe, given its broad rise in different geographical settings. A more likely explanation is the D614G variant’s selective advantage allowing more efficient spread. Dr. Kenney and the Antibody Solutions team applied its molecular modeling platform to dive deeper into the D614G mutation to see if any fresh insights could be yielded from evaluating the effect of the mutation on the virus spike protein.
The team modeled a three-dimensional protein domain of SARS-CoV-2 Spike protein from amino acid (AA) 591 to 710 using an I-TASSER multiple-template threading methodology. The goal was to create a prediction of the secondary structure of the region that contains both the site of mutation and the furin cleavage site that is believed to impart enhanced infectivity onto the virus.
“We created two models and observed a provocative result in which the only significant changes in structure are seen at the furin binding site,” Dr. Kenney said. “Essentially, the D614G mutation changes the orientation of critical residues in the furin cleavage domain and is more favorably aligned within the active site of furin. If furin cleavage is rate-determining in the membrane fusion process, such an increase in protein cleavage would lead to more rapid membrane fusion and entry into cells that serve as hosts for the virus.
“So, our modeling predicts a conformational change in a specific cleavage site induced by the D614G mutation that may reduce the required activation energy and translate into an advantage in infectivity,” he continued.
Implications and next steps
Whether this advantage is conferred by infectivity, immune evasion or pathogenicity—or some combination of these—is yet to be understood. Drs. Kenney and Chen cautioned that their research doesn’t seek to draw specific conclusions about the D614G mutation’s transmissibility, sequelae influence or other broad immunological dynamics relating to the SARS-CoV-2 pandemic. “Our proposed mechanism for how the D614G mutation could lead to increased infectivity certainly does not preclude other mechanisms that could act in concert with the one we’ve proposed here,” Dr. Kenney said.
Dr. Kenney said the team is driving toward a better understanding of the biology of the virus and why the D614G mutation seems to be so central to greater infectivity.
“This is a case where sequencing and molecular modeling working hand-in-hand can help build a greater understanding about the key mechanisms in the pathogenesis of SARS-CoV-2,” he said. “Most mutations either fade from view or, if they stick, don’t have deleterious effects. We believe that the D614G mutation represents one of those instances where there’s a clear case of it increasing the pathogenesis of this virus, which points to the site we identified as an important target for therapeutic antibodies, vaccines and other modalities.
“We hope this research provides an ideal engagement point for researchers working on SARS-CoV-2,” he said. “This wasn’t an ‘obvious find,’ and it took both of our teams working closely together to reveal it. It was a unique insight that we hope will inspire the scientific community to continue exploring. Genuinely, we want to know what we have right and wrong, and we intend to explore therapeutic angles ourselves.”
Dr. Kenney said that in addition to its most recent collaboration with SCT, the company is working on multiple SARS-CoV-2 research initiatives on its own and on behalf of clients pursuing vaccines and therapies for the virus.
“It’s not often that small biotech companies pursue basic research into disease mechanisms,” Dr. Kenney said. “But the serious impact of the SARS-CoV-2 outbreak on our families, friends and community has prompted us to use the tools we have and help find solutions to the COVID-19 pandemic.”
About Antibody Solutions
For more than 25 years, Antibody Solutions has been at the forefront of therapeutic antibody discovery, offering a robust and time-tested antibody discovery platform and utilizing new technologies such as human antibody-producing transgenic animals and molecular modeling software. A privately held contract research organization based in Silicon Valley, Antibody Solutions has helped more than 500 biopharma clients worldwide advance diagnostics, vaccines and therapies with fit-for-purpose antibodies and trusted scientific guidance. Through an acute customer-service focus, the company provides comprehensive project management and technical support in a unique fee-for-service, pay-as-you-go model that helps clients get to market smarter and faster. From its headquarters in Santa Clara, Calif., the company’s broad portfolio of services includes monoclonal and polyclonal antibody development, antigen generation, antibody sequencing, bioreactor technology, pharmacokinetic studies, epitope binning, peptide synthesis, immunoassay development, ligand-binding assay analysis, and support for CAR-T research. For more information, visit antibody.com.
About Single Cell Technology, Inc.
Single Cell Technology (SCT) is a biotechnology company in San Jose, Calif., that has developed a new and revolutionary approach to antibody discovery. Their AbTheneumTM engine can analyze the kinetics and sequence the mRNA from individual antibody secreting cells. Antibodies are rapidly screened against multiple molecules to measure their affinity and specificity, and mRNA from the cognate light and heavy chains are sequenced by Next Generation Sequencing (NGS) technology and correctly paired. SCT integrates advances in multiple disciplines, such as microscale fabrication, molecular biology, precision mechanical engineering, image processing, NGS, and bioinformatics. SCT’s approach substantially decreases the time and investment needed to discover high quality antibodies. Learn more at singlecelltechnology.com.
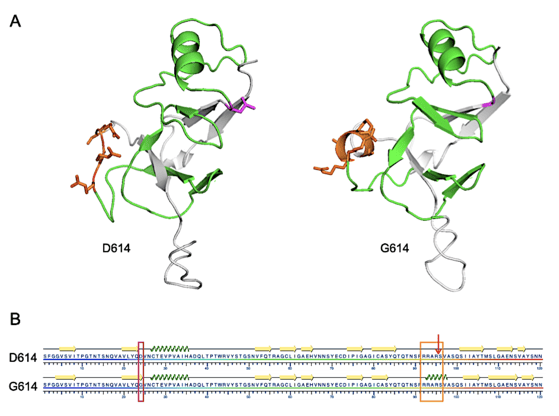
FIGURE 1. Structural representations of D614 and G614 SARS-CoV-2 S protein from S591 through N710.
A. I-TASSER-derived models. The furin cleavage domain 682-RRARS-686 is shown in orange and D614 (left) or G614 (right) are shown in magenta. Residues between 614 and the cleavage domain are shown in green.
B. Results of the KSDSSP secondary structure algorithm. The D614G mutation is highlighted with a magenta box, the furin cleavage domain with an orange box and the cleavage site by the arrow. Beta sheet or alpha helix secondary structures are indicated by yellow arrows or green corkscrew, respectively. From Preprints 2020, 2020050407 (doi: 10.20944/preprints202005.0407.v1).

Author of more than 40 publications, John’s current research interests include new technologies for improving therapeutic antibody discovery, properties of next-generation antibody-like molecules, and best practices for critical reagents used in biologics development.




