5 min read
The Delta Variant Furin Domain Mutation Favors Greater Infectivity
The Delta variant has quickly become the predominant strain of SARS-CoV-2. Based on molecular modeling studies, Antibody Solutions was one of the first to propose that the earliest variant, D614G, was more transmissible by promoting furin processing, a key step in virus infection. Consistent with our modeling results, it was later found that a spike protein construct containing the G614 mutation was more susceptible to S1/S2 cleavage by furin. Subsequent SARS-CoV-2 variants have continued to possess the D614G mutation, including the two variants of greatest concern today: Alpha and Delta.
Infectivity of SARS-CoV-2 is regulated by three sites on the spike protein: a receptor-binding motif (RBM), a subunit (S)1/S2 cleavage site acted upon by the host protease furin, and an S2’ cleavage site acted upon by host protease TMPRSS2. While many mutations have arisen in SARS-CoV-2 variants, we view the RBM and S1/S2 sites in the spike protein as mutational “hot spots” that correlate with improved infectivity of the virus as illustrated below.
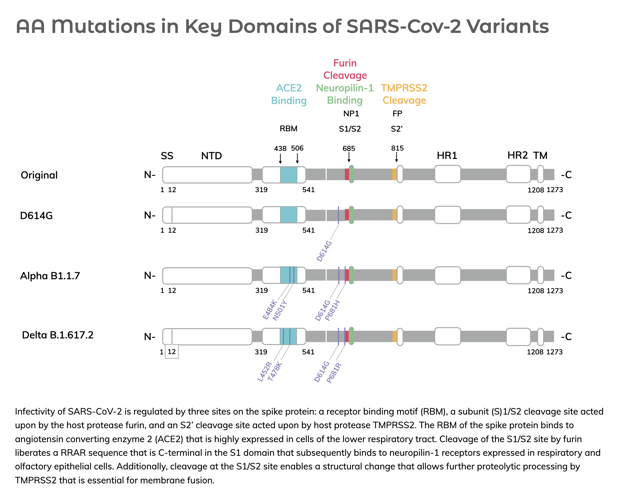
The locations of the key functional domains of SARS-CoV-2 spike protein are shown linearly arranged in the monomer sequence. Mutations occurring in the RBM and proximal to the S1/S2 site are shown in red.
In December, we reported our modeling studies on the spike protein-ACE2 receptor structure of the Alpha variant (aka B.1.1.7). This more transmissible variant has two mutations in the RBM of the spike protein, E484K and N501Y. Using the binding energy of the E484 and N501 construct as the baseline (i.e., ΔE of 0), we calculated the impact of the mutations. The K484 and the Y501 mutations in the RBM of the Alpha variant enhance the stability of the complex with a ΔE of -2.14. Since these mutations energetically favor the interaction of the RBM with the ACE2 receptor, E484K and N501Y would be predicted to be a major driver of higher infectivity for the Alpha variant
The Delta variant has a different set of mutations in the RBM, L452R and T478K, while lacking E484K and N501Y. Our modeling studies of the Delta variant show the L452R and T478K mutations to be destabilizing for the ACE2/spike protein complex with a ΔE of 1.6. In contrast to the Alpha variant, the RBM mutations for the Delta variant do not appear to energetically favor a stronger interaction between the RBM and the ACE2 receptor. What then could account for the increased infectivity of the Delta strain?
Molecular modeling of the ACE2/spike protein RBD complex
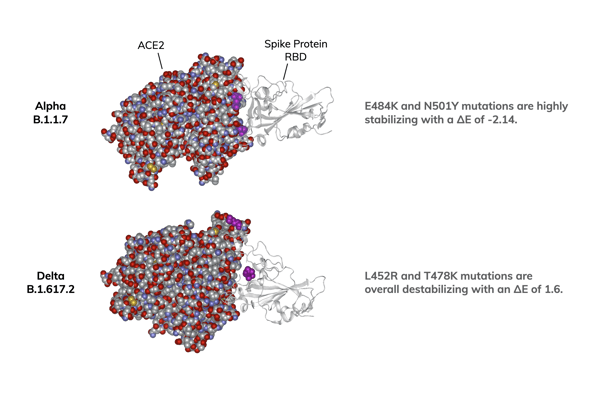
Protein docking analysis was performed by incorporating mutated residues of the variants into models generated from the crystal structure of the original SARS-CoV-2 spike protein RBD bound to ACE2. Energy differences (ΔE) were calculated by minimizing the energy of the variant structure and comparing it to the original structure. Locations of spike protein mutations are shown in magenta.
Furin cleavage of the S1/S2 site is a key step in regulating virus infectivity. Cleavage of the S1/S2 subunit depends upon a polybasic furin cleavage domain, (682)RRAR-S(686), with R682 and R685 being critical residues for catalysis. Both the Alpha and Delta variants have a mutation at P681 in the S1/S2 furin cleavage site. For Alpha, the amino acid proline upstream of the RRAR-S domain is mutated to histidine; for Delta the proline is mutated to arginine. We had shown previously that the D614G mutation arranges the R1 arginine (R685) and R4 arginine (R682) with better proximity to their respective binding pockets of furin.
We compared models of region 590-710 of the original, D614G, Alpha, and Delta SARS-CoV-2 spike proteins. The distance of the critical R4 arginine epsilon atom to the oxygen of the serine at the cleavage site was calculated. R4 to S distances ranged from 12.3 to 6.2 Å in the four models. The Delta variant has the shortest distance of 6.2 Å from R4 to S; additionally, it contains an added arginine at position R5 with a distance to S of 8.3 Å. Both the closer orientation of the R4 arginine and additional R5 arginine proximal to the catalytic site would be predicted to more effectively promote furin cleavage.
Progression of amino acid mutations in the S1/S2 site
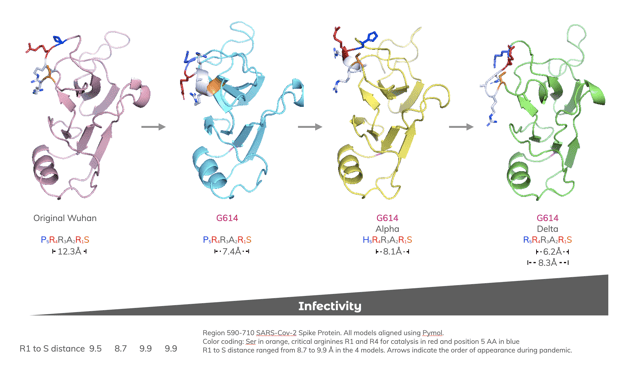
Region 590-710 SARS-CoV-2 spike proteins. All models aligned using Pymol. Color coding: Ser in orange, critical arginines R1 and R4 for catalysis in red and position 5 amino acid in blue. Distances in angstroms (Å) between the epsilon atom of the arginine in the R4 or R5 positions and the side chain oxygen of the Serine (S) at the cleavage site are indicated. Arrows indicate the order of appearance during pandemic.
Consistent with our modeling results, Liu, et al have recently published results showing that the Delta variant virus has a higher replication fitness than the Alpha variant. They ascribed the higher fitness of Delta to more efficient furin cleavage of the S1/S2 site compared to Alpha or wild-type virus. Furthermore, Liu, et al. observed the Alpha spike protein to have a significantly higher affinity for ACE2 compared to the Delta spike, as predicted by our modeling.
Taken together, these observations suggest that the higher infectivity of the Delta variant is not associated with the RBD mutations as is the case for Alpha. Instead, they point to the P681R spike mutation as the main driver for enhanced infectivity of the Delta variant. More efficient cleavage by furin of the Delta S1/S2 domain (i.e., RRRAR-S) could impact two mechanisms of viral infection: enhanced protein processing needed for membrane fusion, and increased expression of the RRAR C-terminal NRP1 binding site.
During the pandemic, the receptor binding domain of the SARS-CoV-2 spike protein has been the main target for therapeutic antibodies that can be used to treat COVID-19. The rise and predominance of the Delta variant that contains a mutation favoring S1/S2 cleavage by furin points to the S1/S2 domain as an additional target for antibody discovery. We’ll continue to publish additional findings from our SARS-CoV-2 research, and we encourage you to please let us know what you’re discovering in your studies.

Author of more than 40 publications, John’s current research interests include new technologies for improving therapeutic antibody discovery, properties of next-generation antibody-like molecules, and best practices for critical reagents used in biologics development.





